Get an overview of how Dextramer® reagents can assist with vaccine development.
Vaccines are a promising strategy to treat cancer by coaxing the immune system to recognize and eliminate tumor cells. A vaccine’s efficacy in eliciting a safe and predictable outcome builds on a clear understanding of the behavior of immune cells in response to antigens. Thus, characterizing and monitoring antigen-specific immune cells is key to cancer vaccine development.
In this blog post, we briefly review existing cancer vaccines and explore how MHC multimers for the detection of antigen-specific immune cells, like Dextramer® reagents, can help to determine why some patients respond to vaccines while others don’t, when a vaccine’s effect might wane, and how vaccines can be used to improve treatment outcomes.
Cancer vaccines can be classified based on the source, nature, and delivery of the antigens used¹ (Figure 1). Anonymous antigen vaccines do not require antigen identification. Instead, tumor cells or lysates are co-localized with antigen-presenting cells (APC) either in the lab or at the tumor site (in vivo) to induce immune activation. For example, T-VEC is an in situ oncolytic viral vaccine that is delivered intra-lesionally.² These vaccines capture the diversity of tumor antigens (including unique and mutated antigens) but have failed to elicit durable immune responses.
Most vaccines in development target pre-defined antigens that fall into two categories: Shared antigens and private neoantigens.
Pre-defined antigen vaccines are delivered to patients either as a peptide or, more frequently, as a peptide-encoding nucleic acid vaccine. The objective is to induce APCs to present cancer epitopes on the cell surface in a pMHC complex in order to elicit a T-cell response. Identifying and selecting the multiple epitopes to include in a cancer vaccine is a complex process.³ Effective immune monitoring is, therefore, crucial in vaccine development to assess that the vaccine activates and expands T cells with the intended tumor specificities.³
Mechanism of action of cancer vaccines
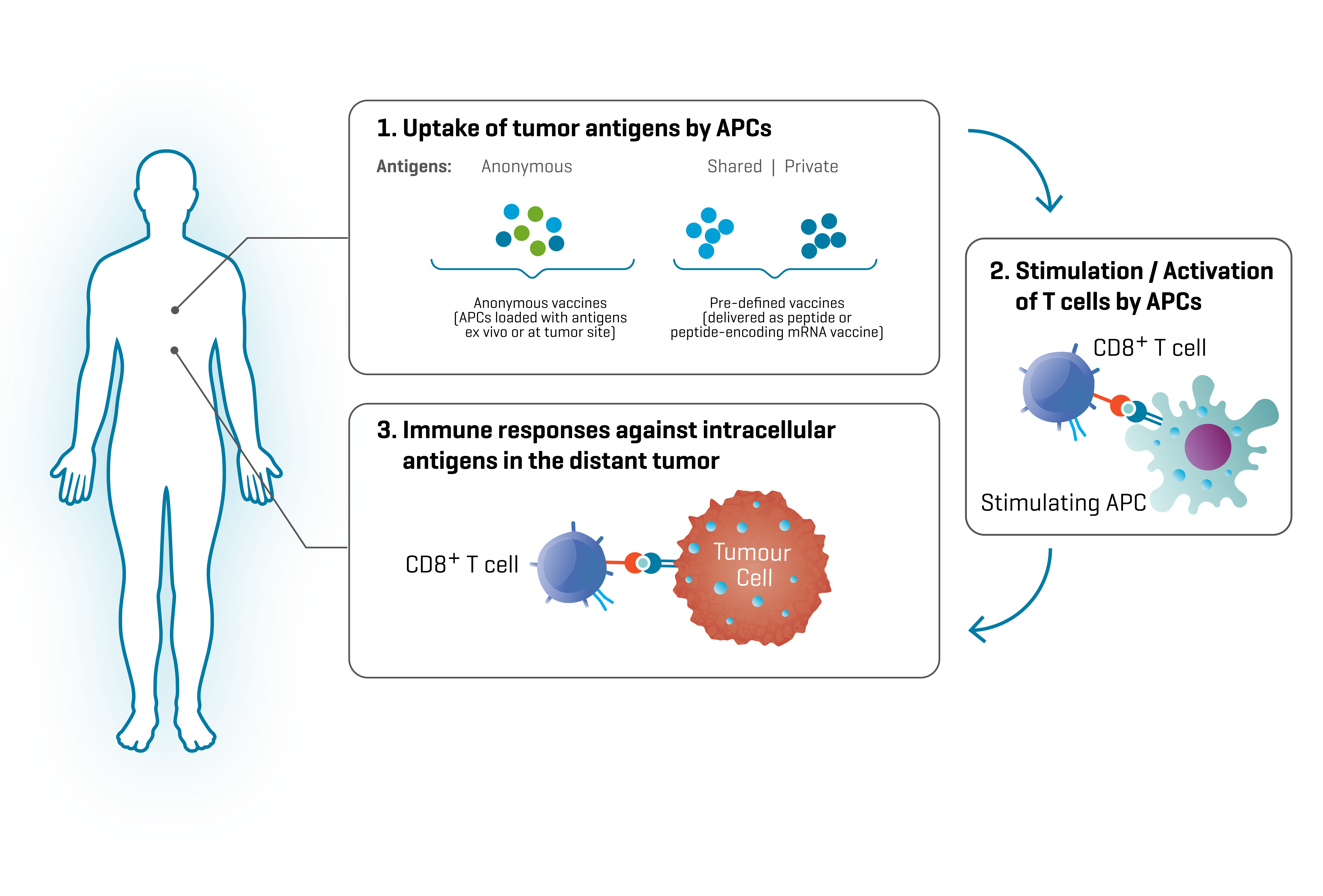
Figure 1. Cancer vaccines may be broadly divided into Anonymous cancer vaccines and those that target Pre-defined Antigens – either shared or private neoantigens. In the case of anonymous cancer vaccines, APCs are loaded with antigens either in the lab or at the tumor site (in vivo). Pre-defined cancer vaccines require the identification of antigens that are either shared or private to a certain patient, and are delivered as a peptide or peptide-encoding nucleic acid vaccine. The antigens are taken up by APCs, presented on the cell surface in a pMHC complex, resulting in stimulation/activation of T cells, and ultimately inducing an immune response in the tumor against the intracellular antigens. Figure inspired by Lin et al.¹
Given the intention to prompt a cellular assault on malignancies, understanding the specificity, phenotype, activity, and persistence of the tumor-targeting immune cells is fundamental to designing and developing more effective and accessible cancer vaccines. MHC multimers, like MHC Dextramer® reagents, are used to monitor the effects of vaccines at various stages of development and testing. The following are a few examples.
Some patients respond to cancer vaccines; others do not. Why? How do responders differ from non-responders? In a pilot trial assessing a novel vaccine targeting H3.3K27M, an antigen shared among patients with diffuse midline gliomas, antigen-specific CD8+ T cells were enumerated in patients after multidose immunization. Staining with H3.3K27M Dextramer® reagents and analysis by flow cytometry and mass spectrometry showed that the prolonged overall survival of responders compared to non-responders (16.3 vs. 9.9 months) was associated with an expansion of H3.3K27M-reactive T cells.
Figure 2 illustrates the workflow for staining of patient samples with MHC Dextramer® reagents and analysis of antigen-specific T cells by flow cytometry. Comparing the frequency, phenotype, and function of T cells specific to the vaccine antigen can reveal mechanisms of action and identify biomarkers of response and prognosis.
Monitoring Tumor-Specific T Cells with MHC Dextramer®
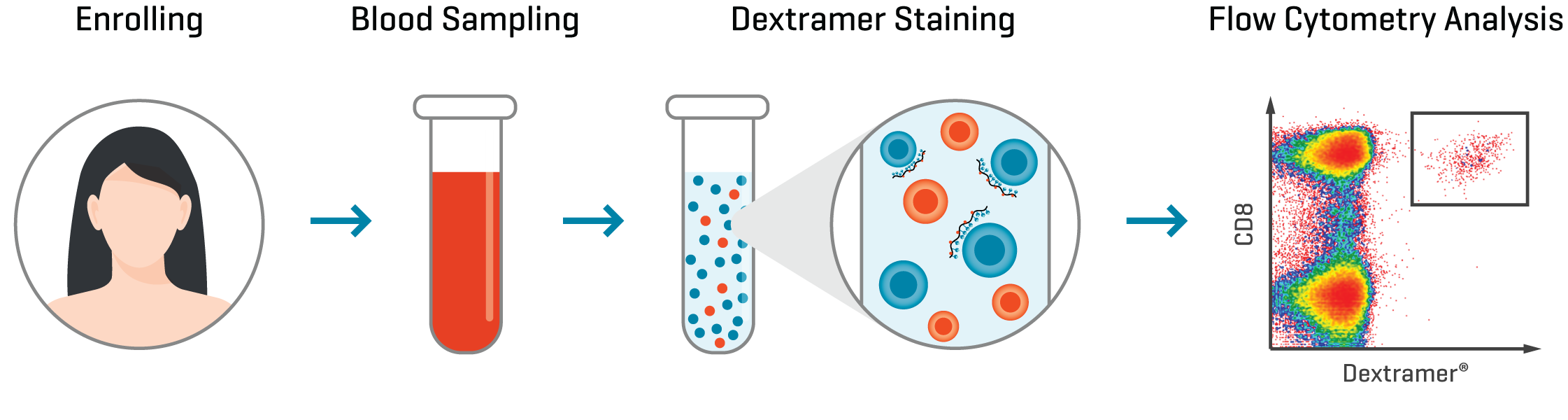
Figure 2. Antigen-specific MHC Dextramer® reagents can be used to monitor the presence, frequency, and phenotype of tumor-specific T cells in patients following vaccination using flow cytometry.
Longitudinal characterization of the immune response to vaccination can paint a long-term picture of the persistence of protection afforded by a cancer vaccine. The FixVac mRNA vaccine developed by BioNTech is an example of a shared antigen vaccine for the treatment of melanoma. The mRNA encodes four non-mutated melanoma-associated antigens aimed at triggering a strong and precise immune response in patients with advanced melanoma. The mRNA is formulated in lipid nanoparticles designed to deliver their payload specifically to antigen presenting dendritic cells.
A dose escalation phase I clinical trial in patients with advanced melanoma demonstrated that the FixVac vaccine stimulated strong cytotoxic immunity in over 75% of the vaccinated patients. Using antigen-specific MHC Dextramer® reagents, the researchers monitored CD4+ and CD8+ T-cell dynamics for a year after immunization. They established that responsive patients showed polyclonal T-cell responses against at least one of the vaccine antigens and that the induced CD8+ T cells had an effector memory phenotype that persisted throughout the study period of 300 days, during which patients received multiple vaccinations.
Figure 3 illustrates how longitudinal monitoring of tumor-specific T cells can be helpful as part of immune monitoring following vaccination.
Longitudinal Monitoring of Tumor-Specific T Cells
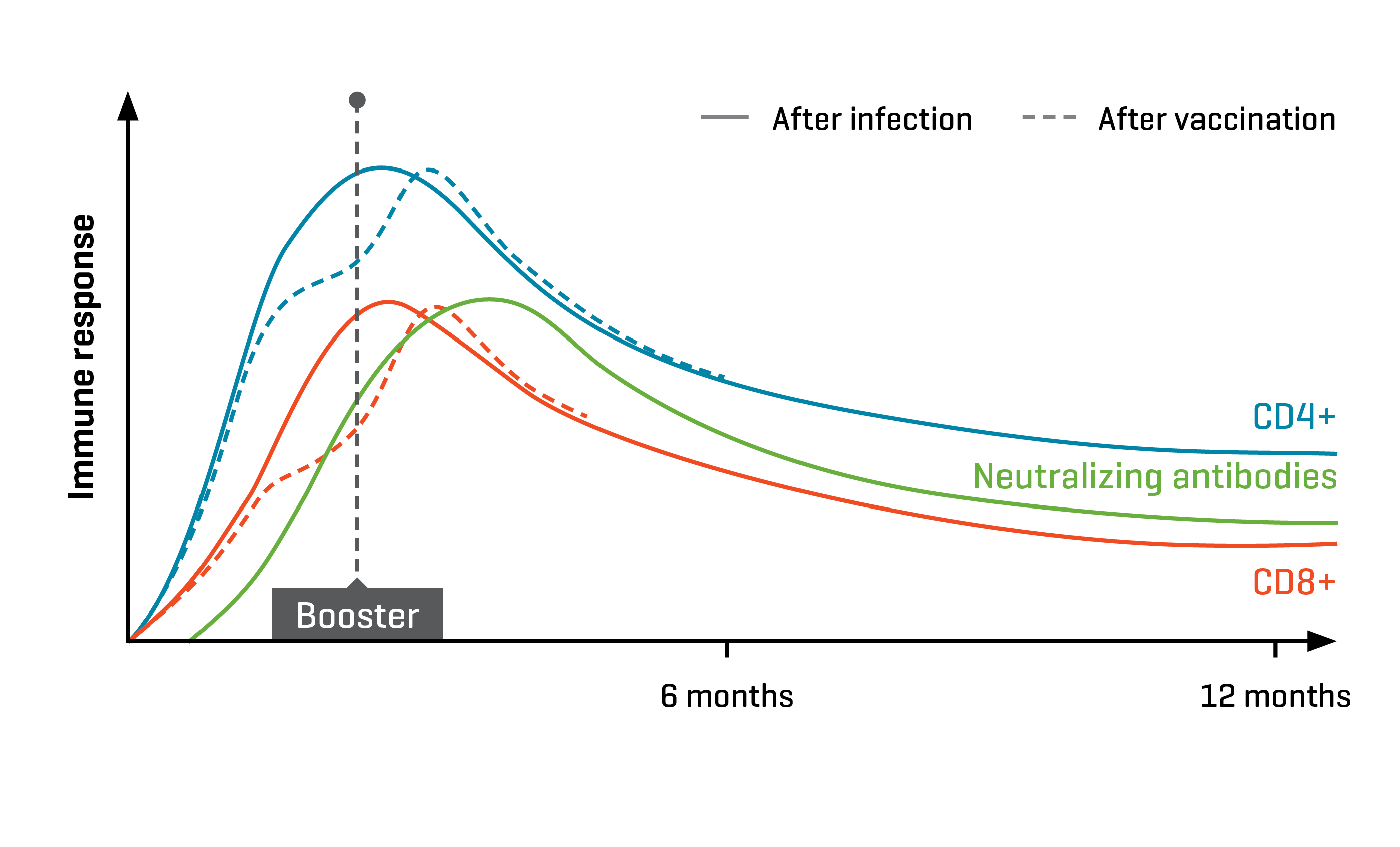
Figure 3. Schematic diagram illustrating how antigen-specific MHC Dextramer® reagents can be used to monitor the presence and frequency of tumor-specific T cells in patients participating in clinical trials as part of long-term immune monitoring after vaccination. This may help to understand the durability of the immune response and the impact of booster vaccinations.
Examining novel vaccine modalities – new delivery systems, combination therapies – benefits from deep immune monitoring to inform strategies that improve efficacy or correlate immune cell action with clinical outcomes. For example, researchers have characterized the response of tumor-infiltrating leukocytes (TILs) in mice to a vaccine delivering a synthetic long peptide as an antigen in liposomes. Using Dextramer® reagents to capture antigen-specific CD8+ TILs, they demonstrated that the vaccine-expanded TILs exhibited a strong exhaustion signature compared to bulk TILs. Thus, they suggested combining the liposome vaccine with a checkpoint inhibitor to prevent exhaustion.
In another example, a plasmacytoid dendritic cell line was used to boost the efficacy of a checkpoint inhibitor to treat metastatic melanoma. Checkpoint inhibitors are effective only when antitumor immunity is already present in a patient. Dendritic cells are powerful immune inducers. So, the researchers tested if an allogeneic plasmacytoid dendritic cell line loaded with four shared melanoma antigens could prime and expand an antitumor response in melanoma patients. That effect would then potentiate subsequent treatment with checkpoint inhibitors. They used MHC I Dextramer® staining to demonstrate that the vaccine increased the frequency of CD8+ T cells specific to the loaded antigens in vaccinated melanoma patients and that this expansion was accompanied by a switch from naïve to memory phenotype. The research group obtained similar results with this vaccine strategy in non-small cell lung cancer patients.⁴
A relatively unexplored context in the development of cancer vaccines is the interplay between different immune cell types that become activated by the vaccine’s antigens. It’s common to talk about T-cell responses to cancer vaccines; however, B cells can also contribute to anti-tumor mechanisms as immune regulators, antigen-presenting cells,⁵ and hubs of humoral immunity.⁶
An in-depth functional characterization of H3K27M vaccination for diffuse midline gliomas revealed that the resulting response of memory CD4+ T cells led to the activation of H3K27M-specific B cells in cerebrospinal fluid. The authors suggested that the presence of these tumor-reactive B cells may serve as antigen-presenting support to maintain the function of the memory CD4+ T cells.⁷
Klickmer® is a flexible technology for investigating and characterizing antigen-specific B cells. Klickmer® can be loaded with your choice of biotinylated antigen(s) to create tailored multimers that efficiently bind B cells (Figure 4).
B Cells Can Also Contribute to Anti-Tumor Mechanisms
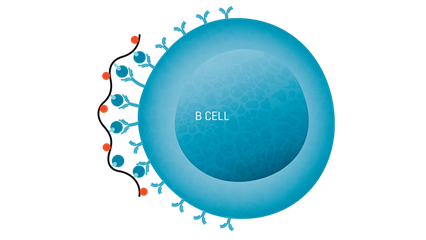
Figure 4. The role of B cells in anti-tumor mechanisms can be investigated using Klickmer® technology to detect and quantify antigen-specific B cells. MHC Dextramer® and Klickmer® reagents can be used together for the simultaneous analysis of antigen-specific T and B cells in a single sample.
Progress in cancer vaccines has been difficult.⁸ Tumor antigens are hard to identify, and tumor cells often develop mechanisms to evade or suppress the immune system. For example, surface expression of specific epitopes being targeted by the immune system can be down-regulated, either through down-regulation of the parent protein or by changing its processing and presentation. But the field is evolving rapidly (Figure 5) and the landmark success of mRNA/lipid nanoparticle SARS-CoV2 vaccines gives reason for optimism that this drug modality can yield breakthroughs in cancer therapy as well.
The Rise in Cancer Vaccine Publications
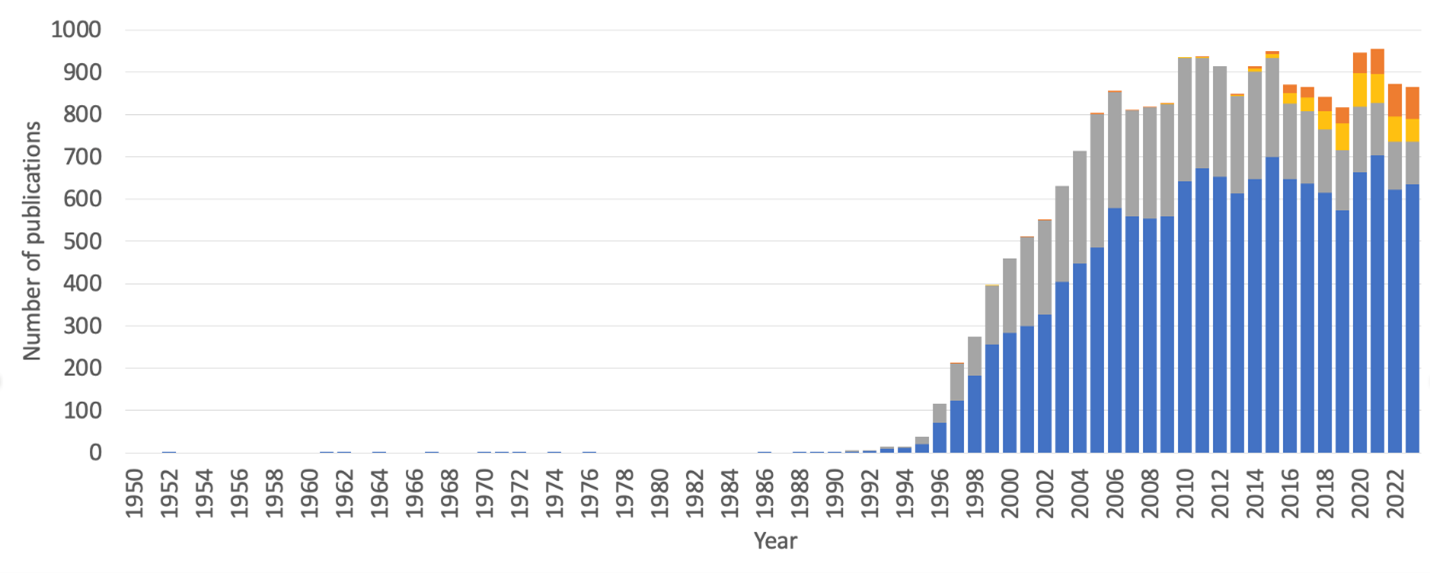
Figure 5. Number of publications per year retrieved in a PubMed search using the MeSH term “cancer vaccines”. Each bar represents all publications in a year. The gray portion highlights papers that additionally included the keywords “tumor-associated antigen” or “tumor-specific antigen”. The orange portion consists of papers that additionally include the keywords “neoepitope” or “neoantigen.” Finally, the yellow portion encompasses overlapping papers using any combination of the first two keywords with the second two.
Numerous clinical trials are underway examining new vaccine strategies often in combination with immune checkpoint blockade therapy and some with promising initial results. With the right design, cancer vaccines can potentially induce a long-lasting and tumor-specific immune memory that can prevent tumor recurrence and metastasis.
Extensive immune monitoring is critical to cancer vaccine development and interpretation of clinical trial results. This will involve not only enumeration of antigen specific T-cells, but also characterization of T-cell phenotypes to assess their anti-tumor activity.
Combining multiplex staining of T cells with barcoded MHC multimers and single-cell multi-omics technology is an approach that enables identification of antigen-specific clonotypic amplification coupled with information about T-cell phenotype and function (e.g., differentiation, activation/exhaustion, cytokine expression; Figure 6). Monitoring changes in the clonal diversity of immune cells over time may provide valuable information about which clonotypes are dominant, expanding, or contracting. A single dominant clonotype could predispose to relapse and vaccine escape, while a broad clonotype distribution may prevent that.
Monitoring Clonal Diversity of Tumor-Specific T Cells using dCODE Dextramer®
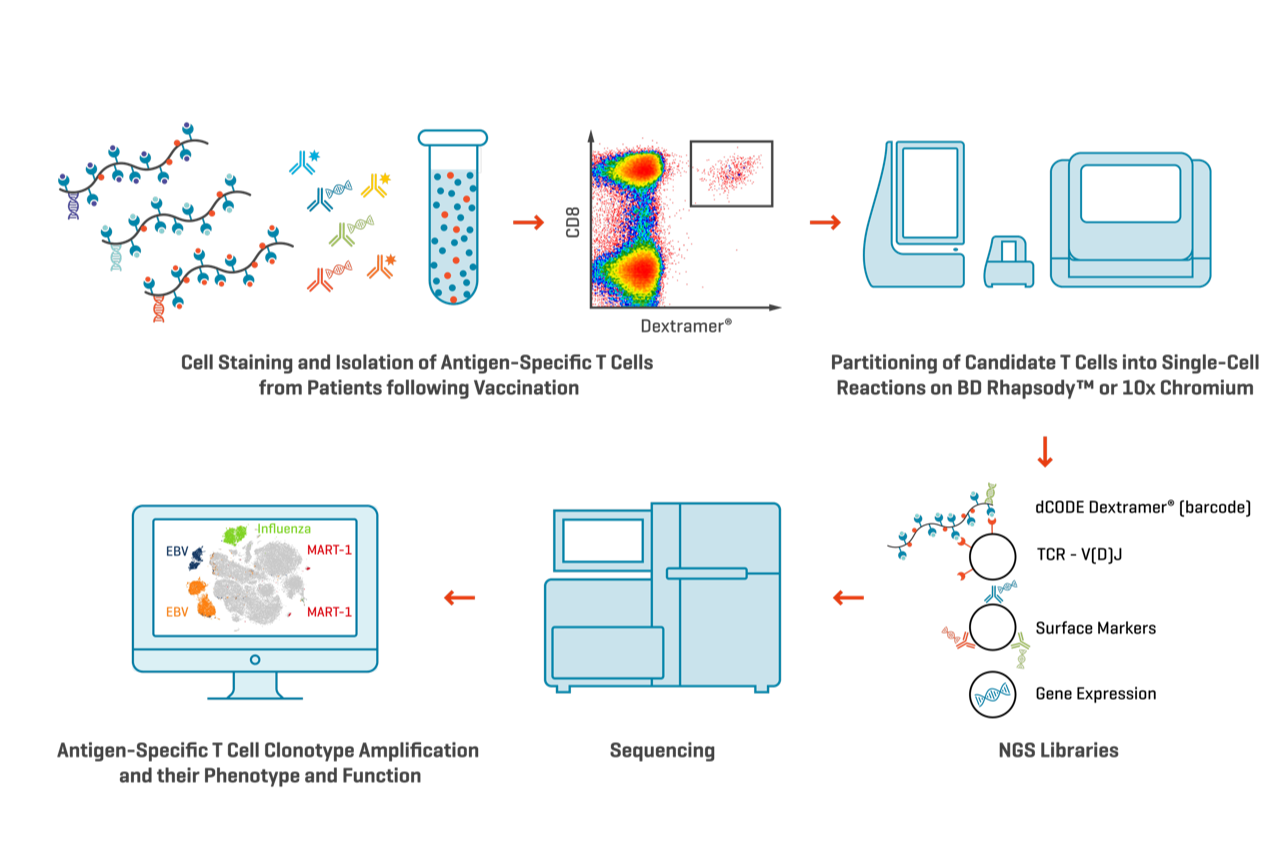
Figure 6. Multiple dCODE Dextramer® reagents, each with different specificities targeting a range of antigen-specific T cells, may be pooled together for multiplexed single-cell multi-omics analysis. The resulting TCR sequencing data will provide information on T cell clonal diversity and distribution. dCODE Dextramer® and dCODE Klickmer® reagents may be used together for the simultaneous analysis of antigen-specific T and B cells in a single sample.
But are we asking the right questions by focusing solely on T cells? As illustrated in the Boschert et al. study that explored T- and B cell responses,⁷ a better understanding of vaccine effects might come from a more nuanced characterization of the combined activity of multiple immune cell types.
As we consider next-generation vaccine technologies that build on novel vaccine modalities (Table 1), it stands to reason that we need to leverage the full armory of the cellular immune response.⁹
The tools to do so are available.
Talk to us to learn how Immudex reagents can help.
Vaccine modality |
Target antigens |
Model |
Insights from experiments using Dextramer® Reagents |
Citation |
|
Two viruses expressing 4 shared antigens in prime-boost regime |
5T4, PSA, PAP, STEAP1 |
Prostate cancer (mouse model) | Exposure to one virus-based vaccine followed by booster with the second induced a large surge in functional CD8+ T cells with high in vivo killing activity. | Vardeu et al. 2022 |
|
Adenovirus-inspired virus-like particle displaying a large tumor-associated antigen |
OVA |
Melanoma (mouse model) |
Percent OT-I-specific CD8+ T cells in spleen was not significantly increased despite near complete clearance of tumors (OT-I is an MHC I-restricted epitope of OVA). Percent may have been higher in tumors, but material was too limited to test. The authors suggest that a multi-specific immune response may account for the strong tumor clearance. |
|
|
Great Apes adenovirus-vectored vaccine with 7 neopitopes fused together as a single antigen. Combined with PD-1 blocking. |
Adpgk, Reps1, Cpne1 |
Colorectal cancer |
Flow cytometry and single-cell RNA-Seq of Adpgk-specific CD8+ T cells revealed a dynamic cell population expansion with stem-like T cells in draining lymph nodes, memory T cell precursors in spleen, and effector T cells in the tumor bed. TCR sequencing showed greater inter- and intraclonal heterogeneity of Adpgk-specific CD8+ T cells in the combination treatment compared to PD-1 monotherapy. |
|
|
Bacteriophage-based nanoparticles delivering NY-ESO-1 antigen and α-GalCer adjuvant |
NY-ESO-1 |
Transgenic HHK mice |
Higher numbers of NY-ESO-1-specific CD8+ T cells were detected in the spleens of HHK mice injected with the vaccine compared to PBS, the phage with only NY-ESO-1, or NY-ESO-1 alone. |
|
|
Lipid-nanoparticle encapsulated mRNA vaccine |
HPV E7 |
Oropharyngeal cancer |
HPV E7-specific CD8+ T cells activated by vaccination were more effector-like and antigen-experienced than non-specific cytotoxic T cells. Their expansion was greater in spleen and blood of mice vaccinated intravenously compared to subcutaneous administration. Their abundance in spleen, blood, and the tumor microenvironment was also inversely correlated with tumor volume. |
|
|
Senescent cancer cell vaccine |
None |
BALB/c mice injected with senescent colon carcinoma cells (CT26) |
Mice injected with senescent CT26 cells had significantly higher numbers of AH1-specific CD8+ T cells in draining lymph nodes (AH1 is expressed by many BALB/c -derived tumors). Vaccination with the senescent CT26 cells prevented tumor formation when mice were injected with proliferating CT26 cells. |
¹ Lin, M.J. et al. 2022. Nature Cancer 3: 911. https://doi.org/10.1038/s43018-022-00418-6
² Ferrucci, P.F. et al. 2021. Cancers (Basel) 13: 1383. https://doi.org/10.3390/cancers13061383
³ Ott, P.A. et al. 2017. Nature 547: 217. https://doi.org/10.1038/nature22991
⁴ Hannani, D. et al. 2023. Int. J. Mol. Sci. 24: 1897. https://doi.org/10.3390/ijms24031897
⁵ Wennhold, K. et al. 2019. Transfus Med Hemother. 46: 36. https://doi.org/10.1159/000496166
⁶ Kaumaya, P.T.P. 2020. Future Oncol. 16: 1767. https://doi.org/10.2217/fon-2020-0224
⁷ Boschert, T. et al. 2024. Sci Adv 10: eadi9091. https://doi.org/10.1126/sciadv.adi9091
⁸ Fukuhara, H. et al. 2016. Cancer Sci. 107: 1373–1379. https://doi.org/10.1111/cas.13027
⁹ Li, W-H. et al. 2022. Acc. Chem. Res. 55: 2660. https://doi.org/10.1021/acs.accounts.2c00360
Get an overview of how Dextramer® reagents can assist with vaccine development.
Explore how Dextramer® technology can uncover antigen-specific immune responses and transform vaccine development.
Sensitively quantify target immune cells by flow cytometry with a strong signal-to-noise ratio.
Gold standard MHC multimers for analysis of antigen-specific T cells by NGS or single-cell multi-omics.
Fill in the form and one of our dedicated specialists will get in touch with you shortly.