Dive into the range of in silico tools available to predict pMHC binding affinity.
How Do You Validate an Epitope?
Epitope discovery and validation underpin the development of vaccines, diagnostics, and next-generation immunotherapies.
Epitope discovery may involve both experimental methods and computational prediction techniques. For example, the Immune Epitope Database (IEDB) is a comprehensive resource for epitope analysis and prediction, offering tools for MHC binding predictions and access to a vast amount of experimental data.
However, as our understanding of antigenic landscapes deepens, rigorous experimental validation of epitope candidates becomes not just a scientific necessity, but a strategic imperative for translating immunological insights into clinically actionable outcomes.
Screening and validating novel epitopes experimentally can involve a range of approaches, including:
- Tools for prediction of peptide-MHC binding
- Experimental validation of peptide-MHC binding
- pMHC Multimer screening by flow cytometry
- Functional Assays
In this blog, we outline how MHC monomers, MHC multimers and Xynapse™ reagents from Immudex enable epitopes to be validated experimentally.
“pMHC multimer experiments are considered the “gold standard” for validating that a given sequence is a real epitope.”
Epitope Discovery and Validation Workflow
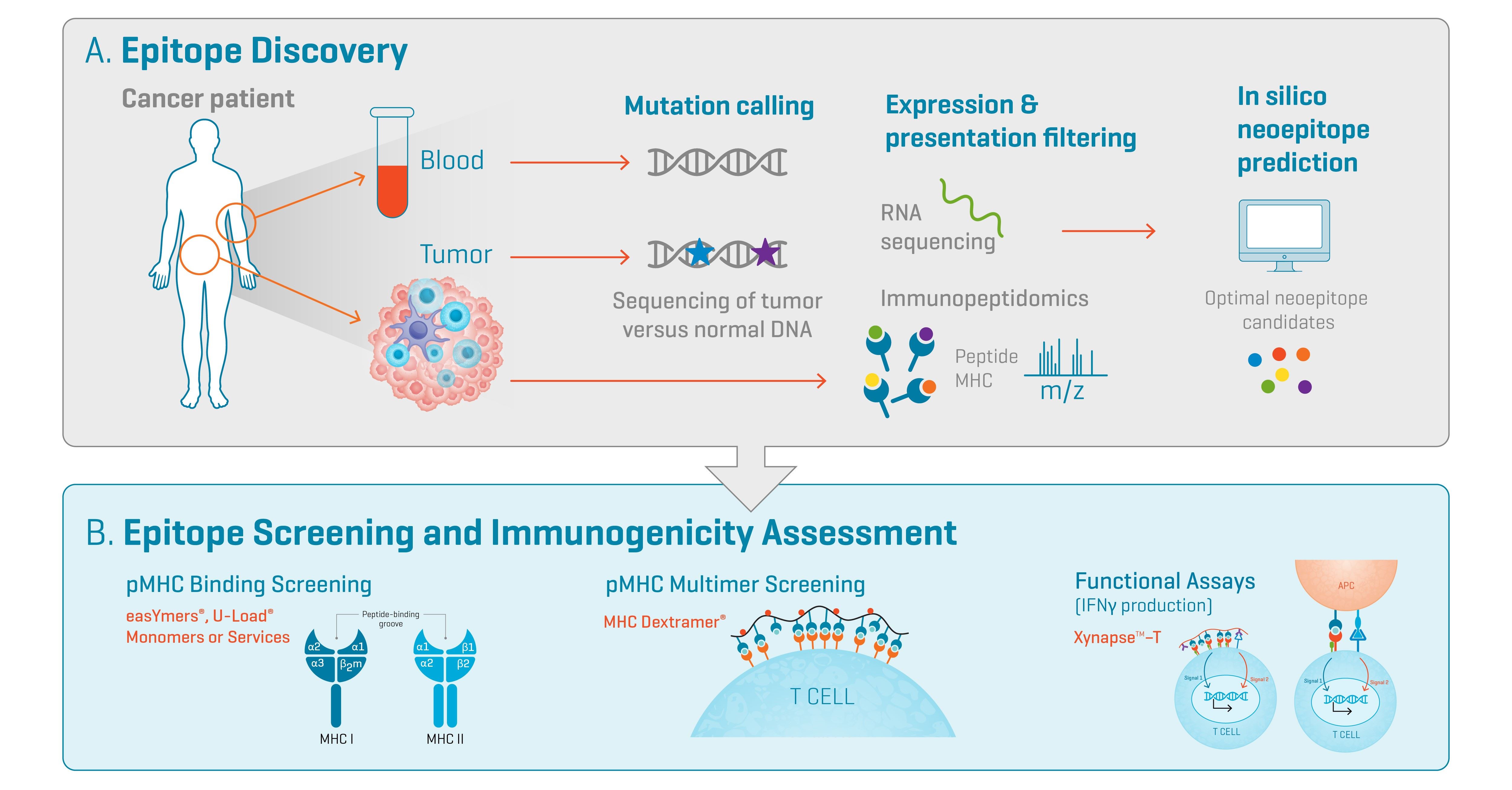
Fig. 1. Overview of tumor neoantigen discovery and validation workflows. A) Epitope Discovery. Non-synonymous mutations are called by whole-exome or whole-genome sequencing of tumor versus germline DNA. These mutations can be filtered by expression (RNA sequencing) and/or presentation (immunopeptidomics). In silico algorithms are used to identify optimal neoepitope candidates. B) Epitope Validation. The selected neoepitope candidates can be validated by assessing pMHC binding experimentally, by pMHC multimer screening by flow cytometry and by functional assays where T cells are co-incubated with APCs or non-cellular antigen-presenting scaffolds such as Xynapse™-T. Figure adapted from Arnaud et al.¹
Validation of pMHC Binding in Your Own Lab
If you would like to validate in your own lab that a given pMHC complex can be folded, peptide-receptive easYmers® MHC I and U-Load® MHC II monomers can be used to generate specific MHC-peptide complexes by loading your peptide of choice. easYmers® also offer the option of performing a flow cytometry-based quality control assay to determine the peptide loading efficiency relative to a positive and negative control, enabling you to evaluate the formation of MHC I monomers.
pMHC Binding Screening Service
At Immudex we also offer a pMHC Binding Screening Service which combines an accurate prediction with experimental validation to ensure reliable identification of strong binders.
We evaluate the binding affinity of peptides to MHC I and II alleles through small-scale refolding of the pMHC complexes of choice. The assays we use assess pMHC binding affinity relative to a positive control peptide.
Find out how this service helps you to assess the immunogenicity of your peptides.
pMHC Multimer Screening by Flow Cytometry
pMHC multimer experiments are considered the “gold standard” for validating that a given sequence is a real epitope. Immudex offers a range of ready-to-use MHC Dextramer® and loadable U-Load Dextramer® reagents for the direct detection and quantification of antigen-specific CD8+ or CD4+ T cells via flow cytometry.
Alternatively, custom MHC Dextramer® reagents can be ordered directly from Immudex, just contact us with your request. We evaluate each unique peptide's ability to bind to the requested MHC allele(s) using in silico approaches.
Functional Assays
In addition, functional assays may be performed – this has been traditionally done by loading candidate peptides into antigen-presenting cells (APCs) and assessing immunogenicity by IFN𝛾 ELISpot assay or by upregulation of activation markers.
Xynapse™ reagents are now available which offer a convenient alternative to stimulation with peptide-pulsed APCs. Xynapse™-T reagents consist of non-cellular antigen-presenting scaffolds that elicit activation of antigen-specific T cells comparable to peptide-pulsed APCs.

References
¹. Arnaud et al. Biotechnologies to tackle the challenge of neoantigen identification. Current Opinion in Biotechnology, 2020. https://doi.org/10.1016/j.copbio.2019.12.014.
Related Resources
Factors Contributing to Immunogenicity
What factors play a role in immunogenicity and pMHC complex binding affinity?
MHC Dextramer®
Explore our range of reagents for validating epitopes by pMHC multimer screening.
Xynapse™-T Reagents
Discover the convenient alternative to stimulation with APCs for antigen-specific T cell activation.
Interested in Epitope Discovery or Validation?
Fill in the form and one of our dedicated specialists will get in touch with you shortly.

